Digital microfluidics
Digital microfluidics (DMF) is a platform for lab-on-a-chip systems that is based upon the manipulation of microdroplets. Droplets are dispensed, moved, stored, mixed, reacted, or analyzed on a platform with a set of insulated electrodes.[1][2] Digital microfluidics can be used together with analytical analysis procedures such as mass spectrometry, colorimetry, electrochemical, and electrochemiluminescense.[1]
Overview
[edit]
In analogy to digital microelectronics, digital microfluidic operations can be combined and reused within hierarchical design structures so that complex procedures (e.g. chemical synthesis or biological assays) can be built up step-by-step. And in contrast to continuous-flow microfluidics, digital microfluidics[3] works much the same way as traditional bench-top protocols, only with much smaller volumes and much higher automation. Thus a wide range of established chemical procedures and protocols can be seamlessly transferred to a nanoliter droplet format. Electrowetting, dielectrophoresis, and immiscible-fluid flows are the three most commonly used principles, which have been used to generate and manipulate microdroplets in a digital microfluidic device.
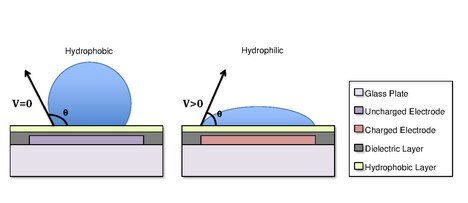
A digital microfluidic (DMF) device set-up depends on the substrates used, the electrodes, the configuration of those electrodes, the use of a dielectric material, the thickness of that dielectric material, the hydrophobic layers, and the applied voltage.[4][5]

A common substrate used in this type of system is glass. Depending if the system is open or closed, there would be either one or two layers of glass. The bottom layer of the device contains a patterned array of individually controllable electrodes.[4] When looking at a closed system, there is usually a continuous ground electrode found through the top layer made usually of indium tin oxide (ITO). The dielectric layer is found around the electrodes in the bottom layer of the device and is important for building up charges and electrical field gradients on the device.[5] A hydrophobic layer is applied to the top layer of the system to decrease the surface energy where the droplet will actually we be in contact with.[5] The applied voltage activates the electrodes and allows changes in the wettability of droplet on the device’s surface. In order to move a droplet, a control voltage is applied to an electrode adjacent to the droplet, and at the same time, the electrode just under the droplet is deactivated. By varying the electric potential along a linear array of electrodes, electrowetting can be used to move droplets along this line of electrodes.[6]
Modifications to this foundation can also be fabricated into the basic design structure. One example of this is the addition of electrochemiluminescence detectors within the indium tin oxide layer (the ground electrode in a closed system) which aid in the detection of luminophores in droplets.[7] In general, different materials may also be used to replace basic components of a DMF system such as the use of PDMS instead of glass for the substrate.[8] Liquid materials can be added, such as oil or another substance, to a closed system to prevent evaporation of materials and decrease surface contamination.[6][9] Also, DMF systems can be compatible with ionic liquid droplets with the use of an oil in a closed device or with the use of a catena (a suspended wire) over an open DMF device.[9]
Digital microfluidics can be light-activated. Optoelectrowetting can be used to transport sessile droplets around a surface containing patterned photoconductors.[10] The photoelectrowetting effect[11] can also be used to achieve droplet transport on a silicon wafer without the necessity of patterned electrodes.[12]
Working principle
[edit]Droplets are formed using the surface tension properties of a liquid. For example, water placed on a hydrophobic surface such as wax paper will form spherical droplets to minimize its contact with the surface.[13] Differences in surface hydrophobicity affect a liquid’s ability to spread and ‘wet’ a surface by changing the contact angle.[14] As the hydrophobicity of a surface increases, the contact angle increases, and the ability of the droplet to wet the surface decreases. The change in contact angle, and therefore wetting, is regulated by the Young-Lippmann equation.[4][9][5]
where is the contact angle with an applied voltage ; is the contact angle with no voltage; is the relative permittivity of the dielectric; is the permittivity of free space; is the liquid/filler media surface tension; is the dielectric thickness.[5]
In some cases, the hydrophobicity of a substrate can be controlled by using electrical fields. This refers to the phenomenon Electrowetting On Dielectric (EWOD).[3][4][5] For example, when no electric field is applied to an electrode, the surface will remain hydrophobic and a liquid droplet will form a more spherical droplet with a greater contact angle. When an electric field is applied, a polarized hydrophilic surface is created. The water droplet then becomes flattened and the contact angle decreases. By controlling the localization of this polarization, we can create an interfacial tension gradient that allows controlled displacement of the droplet across the surface of the DMF device.[6]
Droplet formation
[edit]There are two ways to make new droplets with a digital microfluidic device. Either an existing droplet can be split in two, or a new droplet can be made from a reservoir of material.[15] Both processes are only known to work in closed devices,[9][16] though this often is not a problem as the top plates of DMF devices are typically removable,[17] so an open device can be made temporarily closed should droplet formation be necessary.

From an existing droplet
[edit]A droplet can be split by charging two electrodes on opposite sides of a droplet on an uncharged electrode. In the same way a droplet on an uncharged electrode will move towards an adjacent, charged electrode,[6] this droplet will move towards both active electrodes. Liquid moves to either side, which causes the middle of the droplet to neck.[15] For a droplet of the same size as the electrodes, splitting will occur approximately when , as the neck will be at its thinnest.[15] is the radius of curvature of the menisci at the neck, which is negative for a concave curve, and is the radius of curvature of the menisci at the elongated ends of the droplet. This process is simple and consistently results in two droplets of equal volume.[15][18]
The conventional method[19][15] of splitting an existing droplet by simply turning the splitting electrodes on and off produces new droplets of relatively equal volume. However, the new droplets formed by the conventional method show considerable difference in volume.[20][21] This difference is caused by local perturbations due to the rapid mass transport.[21] Even though the difference is negligible in some applications, it can still pose a problem in applications that are highly sensitive to variations in volume,[22][23] such as immunoassays[24] and DNA amplification.[25] To overcome the limitation of the conventional method, an existing droplet can be split by gradually changing the potential of the electrodes at the splitting region instead of simply switching them on and off.[21] Using this method, a noticeable improvement in droplet volume variation, from around 10% variation in volume to less than 1% variation in volume, has been reported.[21]
From a reservoir
[edit]Creating a new droplet from a reservoir of liquid can be done in a similar fashion to splitting a droplet. In this case, the reservoir remains stationary while a sequence of electrodes are used to draw liquid out of the reservoir. This drawn liquid and the reservoir form a neck of liquid, akin to the neck of a splitting droplet but longer, and the collapsing of this neck forms a dispensed droplet from the drawn liquid.[15][26] In contrast to splitting, though, dispensing droplets in this manner is inconsistent in scale and results. There is no reliable distance liquid will need to be pulled from the reservoir for the neck to collapse, if it even collapses at all.[27] Because this distance varies, the volumes of dispensed droplets will also vary within the same device.[27]
Due to these inconsistencies, alternative techniques for dispensing droplets have been used and proposed, including drawing liquid out of reservoirs in geometries that force a thinner neck,[15][28] using a continuous and replenishable electrowetting channel,[22] and moving reservoirs into corners so as to cut the reservoir down the middle.[18][28] Multiple iterations of the latter can produce droplets of more manageable sizes.
Droplet manipulation
[edit]Droplet merging
[edit]As an existing droplet can be split to form discrete droplets using electrodes (see From an existing droplet),[19][15] droplets can be merged into one droplet by electrodes as well.[29][15] Utilizing the same concept applied for creating new droplets through splitting an existing droplet with electrodes, an aqueous droplet resting on an uncharged electrode can move towards a charged electrode where droplets will join and merge into one droplet.[29][15] However, the merged droplet might not always form a circular shape even after the merging process is over due to surface tension.[15] This problem can be solved by implementing a superhydrophobic surface between the droplets and the electrodes.[29] Oil droplets can be merged in the same way as well, but oil droplets will move towards uncharged electrodes unlike aqueous droplets.[30]
Droplet transportation
[edit]Discrete droplets can be transported in a highly controlled way using an array of electrodes.[31][32][30] In the same way droplets move from an uncharged electrode to a charged electrode, or vice versa, droplets can be continuously transported along the electrodes by sequentially energizing the electrodes.[33][30][15] Since droplet transportation involves an array of electrodes, multiple electrodes can be programmed to selectively apply a voltage to each electrode for a better control over transporting multiple droplets.[33]
Displacement by electrostatic actuation
[edit]Three-dimensional droplet actuation has been made possible by implementing a closed system; this system contains a μL sized droplet in immiscible fluid medium. The droplet and medium are then sandwiched between two electromagnetic plates, creating an EM field between the two plates.[34][35] The purpose of this method is to transfer the droplet from a lower planar surface to an upper parallel planar surface and back down via electrostatic forces.[34][36] The physics behind such particle actuation and perpendicular movement can be understood from early works of N. N. Lebedev and I. P. Skal’skaya.[37] In their research, they attempted to model the Maxwell electrical charge acquired by a perfectly round conducting particle in the presence of a uniform magnetic field caused by a perfectly-conducting and infinitely-stretching surface.[37] Their model helps to predict the Z-direction motion of the microdroplets within the device as it points to the magnitude and direction of forces acting upon a micro droplet. This can be used to help accurately predict and correct for unwanted and uncontrollable particle movement. The model explains why failing to employ dielectric coating on one of the two surfaces causes reversal of charge within the droplet upon contact with each electrode and in turn causes the droplets to uncontrollably bounce of between electrodes.
Digital microfluidics (DMF), has already been readily adapted in many biological fields.[38][39][40] By enabling three-dimensional movement within DMF, the technology can be used even more extensively in biological applications, as it could more accurately mimic 3-D microenvironments. A large benefit of employing this type of method is that it allows for two different environments to be accessible by the droplet, which can be taken advantage of by splitting the microfluidic tasks among the two surfaces. For example, while the lower plane can be used to move droplets, the upper plate can carry out the necessary chemical and/or biological processes.[34] This advantage can be translated into practical experiment protocols in the biological community, such as coupling with DNA amplification.[41][36][42] This also allows for the chip to be smaller, and to give researchers more freedom in designing platforms for microdroplet analysis.[34]
All-terrain droplet actuation (ATDA)
[edit]All-terrain microfluidics is a method used to transport liquid droplets over non-traditional surface types.[43] Unlike traditional microfluidics platform, which are generally restricted to planar and horizontal surfaces, ATDA enables droplet manipulation over curved, non-horizontal, and inverted surfaces.[43] This is made possible by incorporating flexible thin sheets of copper and polyimide into the surface via a rapid prototyping method.[43][44] This device works very well with many liquids, including aqueous buffers, solutions of proteins and DNA, and undiluted bovine serum.[43] ATDA is compatible with silicone oil or pluronic additives, such as F-68, which reduce non-specific absorption and biofouling when dealing with biological fluids such as proteins, biological serums, and DNA.[43][45] A drawback of a setup like this is accelerated droplet evaporation.[43] ATDA is a form of open digital microfluidics, and as such the device needs to be encapsulated in a humidified environment in order to minimize droplet evaporation.[46]
Implementation
[edit]In one of various embodiments of EWOD-based microfluidic biochips, investigated first by Cytonix in 1987 [1] Archived 2020-09-19 at the Wayback Machine and subsequently commercialized by Advanced Liquid Logic, there are two parallel glass plates. The bottom plate contains a patterned array of individually controllable electrodes and the top plate is coated with a continuous grounding electrode. A dielectric insulator coated with a hydrophobic is added to the plates to decrease the wet-ability of the surface and to add capacitance between the droplet and the control electrode. The droplet containing biochemical samples and the filler medium, such as the silicone oil, a fluorinated oil, or air, are sandwiched between the plates and the droplets travel inside the filler medium. In order to move a droplet, a control voltage is applied to an electrode adjacent to the droplet, and at the same time, the electrode just under the droplet is deactivated. By varying the electric potential along a linear array of electrodes, electrowetting can be used to move droplets along this line of electrodes.
Applications
[edit]Laboratory automation
[edit]In research fields such as synthetic biology, where highly iterative experimentation is common, considerable efforts have been made to automate workflows.[47][48][49] Digital microfluidics is often touted as a laboratory automation solution, with a number of advantages over alternative solutions such as pipetting robots and droplet microfluidics.[50][51][52] These stated advantages often include a reduction in the required volume of experimental reagents, a reduction in the likelihood of contamination and cross-contamination, potential improvements in reproducibility, increased throughput, individual droplet addressability, and the ability to integrate with sensor and detector modules to perform end-to-end or even closed loop workflow automation.[50][51][52][53]
Reduced experimental footprint
[edit]One of the core advantages of digital microfluidics, and of microfluidics in general, is the use and actuation of picoliter to microliter scale volumes. Workflows adapted from the bench to a DMF system are miniaturized, meaning working volumes are reduced to fractions of what is normally required for conventional methods. For example, Thaitrong et al. developed a DMF system with a capillary electrophoresis (CE) module with the purpose of automating the process of next generation sequencing (NGS) library characterization. Compared to an Agilent BioAnalyzer (an instrument commonly used to measure sequencing library size distribution), the DMF-CE system consumed ten-fold less sample volume.[54] Reducing volumes for a workflow can be especially beneficial if the reagents are expensive or when manipulating rare samples such as circulating tumor cells and prenatal samples.[52] Miniaturization also means a reduction in waste product volumes.
Reduced probability of contamination
[edit]DMF-based workflows, particularly those using a closed configuration with a top-plate ground electrode, have been shown to be less susceptible to outside contamination compared to some conventional laboratory workflows. This can be attributed to minimal user interaction during automated steps, and the fact that the smaller volumes are less exposed to environmental contaminants than larger volumes which would need to be exposed to open air during mixing. Ruan et al. observed minimal contamination from exogenous nonhuman DNA and no cross-contamination between samples while using their DMF-based digital whole genome sequencing system.[52]
Improved reproducibility
[edit]Overcoming issues of reproducibility has become a topic of growing concern across scientific disciplines.[55] Reproducibility can be especially salient when multiple iterations of the same experimental protocol need to be repeated.[56] Using liquid handling robots that can minimize volume loss between experimental steps are often used to reduce error rates and improve reproducibility. An automated DMF system for CRISPR-Cas9 genome editing was described by Sinha et al, and was used to culture and genetically modify H1299 lung cancer cells. The authors noted that no variation in knockout efficiencies across loci was observed when cells were cultured on the DMF device, whereas cells cultured in well-plates showed variability in upstream loci knockout efficiencies. This reduction in variability was attributed to culturing on a DMF device being more homogenous and reproducible compared with well plate methods.[57]
Increased throughput
[edit]While DMF systems cannot match the same throughput achieved by some liquid handling pipetting robots, or by some droplet-based microfluidic systems, there are still throughput advantages when compared to conventional methods carried out manually.[58]
Individual droplet addressability
[edit]DMF allows for droplet level addressability, meaning individual droplets can be treated as spatially distinct microreactors.[50] This level of droplet control is important for workflows where reactions are sensitive to the order of reagent mixing and incubation times, but where the optimal values of these parameters may still need to be determined. These types of workflows are common in cell-free biology, and Liu et al. were able to demonstrate a proof-of-concept DMF-based strategy for carrying out remote-controlled cell-free protein expression on an OpenDrop chip.[59]
Detector module integration for end-to-end and closed-loop automation
[edit]An often cited advantage DMF platforms have is their potential to integrate with on-chip sensors and off-chip detector modules.[50][59] In theory, real-time and end-point data can be used in conjunction with machine learning methods to automate the process of parameter optimization.
Separation and extraction
[edit]Digital microfluidics can be used for separation and extraction of target analytes. These methods include the use of magnetic particles,[60][61][62][63][64][65][66][67] liquid-liquid extraction,[68] optical tweezers,[69] and hydrodynamic effects.[70]
Magnetic particles
[edit]For magnetic particle separations a droplet of solution containing the analyte of interest is placed on a digital microfluidics electrode array and moved by the changes in the charges of the electrodes. The droplet is moved to an electrode with a magnet on one side of the array with magnetic particles functionalized to bind to the analyte. Then it is moved over the electrode, the magnetic field is removed and the particles are suspended in the droplet. The droplet is swirled on the electrode array to ensure mixing. The magnet is reintroduced and the particles are immobilized and the droplet is moved away. This process is repeated with wash and elution buffers to extract the analyte.[60][61][62][63][64][65][66][67]
Magnetic particles coated with antihuman serum albumin antibodies have been used to isolate human serum albumin, as proof of concept work for immunoprecipitation using digital microfluidics.5 DNA extraction from a whole blood sample has also been performed with digital microfluidics.3 The procedure follows the general methodology as the magnetic particles, but includes pre-treatment on the digital microfluidic platform to lyse the cells prior to DNA extraction.[62]
Liquid-liquid extraction
[edit]Liquid-liquid extractions can be carried out on digital microfluidic device by taking advantage of immiscible liquids.9 Two droplets, one containing the analyte in aqueous phase, and the other an immiscible ionic liquid are present on the electrode array. The two droplets are mixed and the ionic liquid extracts the analyte, and the droplets are easily separable.[68]
Optical tweezers
[edit]Optical tweezers have also been used to separate cells in droplets. Two droplets are mixed on an electrode array, one containing the cells, and the other with nutrients or drugs. The droplets are mixed and then optical tweezers are used to move the cells to one side of the larger droplet before it is split.[71][69] For a more detailed explanation on the underlying principles, see Optical tweezers.
Hydrodynamic separation
[edit]Particles have been applied for use outside of magnetic separation, with hydrodynamic forces to separate particles from the bulk of a droplet.[70] This is performed on electrode arrays with a central electrode and ‘slices’ of electrodes surrounding it. Droplets are added onto the array and swirled in a circular pattern, and the hydrodynamic forces from the swirling cause the particles to aggregate onto the central electrode.[70]
Chemical synthesis
[edit]Digital Microfluidics (DMF) allows for precise manipulation and coordination in small-scale chemical synthesis reactions due to its ability to control micro scale volumes of liquid reagents, allowing for overall less reagent use and waste.[72] This technology can be used in the synthesis compounds such as peptidomimetics and PET tracers.[73][74][75] PET tracers require nanogram quantities and as such, DMF allows for automated and rapid synthesis of tracers with 90-95% efficiency compared to conventional macro-scale techniques.[74][76]
Organic reagents are not commonly used in DMF because they tend to wet the DMF device and cause flooding; however synthesis of organic reagents can be achieved through DMF techniques by carrying the organic reagents through an ionic liquid droplet, thus preventing the organic reagent from flooding the DMF device.[77] Droplets are combined together by inducing opposite charges thus attracting them to each other.[78] This allows for automated mixing of droplets. Mixing of droplets are also used to deposit MOF crystals for printing by delivering reagents into wells and evaporating the solutions for crystal deposition.[79] This method of MOF crystal deposition is relatively cheap and does not require extensive robotic equipment.[79]
Chemical synthesis using digital microfluidics (DMF) has been applied to many noteworthy biological reactions. These include polymerase chain reaction (PCR), as well as the formation of DNA and peptides.[77][80] Reduction, alkylation, and enzymatic digestion have also shown robustness and reproducibility utilizing DMF, indicating potential in the synthesis and manipulation of proteomics.[81] Spectra obtained from the products of these reactions are often identical to their library spectra, while only utilizing a small fraction of bench-scale reactants.[73] Thus, conducting these syntheses on the microscale has the benefit of limiting money spent on purchasing reagents and waste products produced while yielding desirable experimental results. However, numerous challenges need to be overcome to push these reactions to completion through DMF. There have been reports of reduced efficiency in chemical reactions as compared to bench-scale versions of the same syntheses, as lower product yields have been observed.[80] Furthermore, since picoliter and nanoliter size samples must be analyzed, any instrument used in analysis needs to be high in sensitivity. In addition, system setup is often difficult due to extensive amounts of wiring and pumps that are required to operate microchannels and reservoirs.[80] Finally, samples are often subject to solvent evaporation which leads to changes in volume and concentration of reactants, and in some cases reactions to not go to completion.[82]
The composition and purity of molecules synthesized by DMF are often determined utilizing classic analytical techniques. Nuclear magnetic resonance (NMR) spectroscopy has been successfully applied to analyze corresponding intermediates, products, and reaction kinetics.[73][83] A potential issue that arises through the use of NMR is low mass sensitivity, however this can be corrected for by employing microcoils that assist in distinguishing molecules of differing masses.[73] This is necessary since the signal-to-noise ratio of sample sizes in the microliter to nanoliter range is dramatically reduced compared to bench-scale sample sizes, and microcoils have been shown to resolve this issue.[84] Mass spectrometry (MS) and high-performance liquid chromatography (HPLC) have also been used to overcome this challenge.[77][73] Although MS is an attractive analytical technique for distinguishing the products of reactions accomplished through DMF, it poses its own weaknesses. Matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) MS have recently been paired with analyzing microfluidic chemical reactions. However, crystallization and dilution associated with these methods often leads to unfavorable side effects, such as sample loss and side reactions occurring.[17] The use of MS in DMF is discussed in more detail in a later section.
Cell culture
[edit]Connecting the DMF chip to use in the field or world-to-chip interfaces have been accomplished by means of manual pumps and reservoirs which deliver microbes, cells, and media to the device.[85] The lack of extensive pumps and valves allow for elaborate multi step applications involving cells performed in a simple and compact system.[58] In one application, microbial cultures have been transferred onto the chip and allowed to grow with the use of sterile procedures and temperature required for microbial incubation. To validate that this was a viable space for microbial growth, a transformation assay was carried out in the device.[85] This involves exposing E.coli to a vector and heat shocking the bacteria until they take up the DNA. This is then followed by running a DNA gel to assure that the wanted vector was taken up by the bacteria. This study found that the DNA indeed was taken up by the bacteria and expressed as predicted.
Human cells have also been manipulated in Digital Microfluidic Immunocytochemistry in Single Cells (DISC) where DMF platforms were used to culture and use antibodies to label phosphorylated proteins in the cell.[86] Cultured cells are then removed and taken off chip for screening. Another technique synthesizes hydrogels within DMF platforms. This process uses electrodes to deliver reagents to produce the hydrogel, and delivery of cell culture reagents for absorption into the gel.[75][45] The hydrogels are an improvement over 2D cell culture because 3D cell culture have increased cell-cell interactions and cel-extracellular matrix interactions.[45] Spherical cell cultures are another method developed around the ability of DMF to deliver droplets to cells. Application of an electric potential allows for automation of droplet transfer directly to the hanging cell culture.[75]][87] This is beneficial as 3 dimensional cell culture and spheroids better mimic in vivo tissue by allowing for more biologically relevant cultures that have cells growing in an extracellular matrix similarly resembling that in the human body.[87] Another use of DMF platforms in cell culture is its ability to conduct in vitro cell-free cloning using single molecule PCR inside droplets.[88] PCR amplified products are then validated by transfection into yeast cells and a Western blot protein identification.[88]
Problems arising from cell culture applications using DMF include protein adsorption to the device floor, and cytotoxicity to cells. To prevent adsorption of protein to the platform's floor, a surfactant stabilized Silicon oil or hexane was used to coat the surface of the device, and droplets were manipulated atop of the oil or hexane.[86] Hexane was later rapidly evaporated from cultures to prevent a toxic effect on cell cultures.[89] Another approach to solve protein adhesion is the addition of Pluronic additives to droplets in the device.[90] Pluronic additives are generally not cytotoxic but some have been shown to be harmful to cell cultures.[46]
Bio-compatibility of device set up is important for biological analyses. Along with finding Pluronic additives that are not cytotoxic, creating a device whose voltage and disruptive movement would not affect cell viability was accomplished. Through the readout of live/dead assays it was shown that neither voltage required to move droplets, nor the motion of moving cultures affected cell viability.[46]
Biological extraction
[edit]Biological separations usually involve low concentration high volume samples. This can pose an issue for digital microfluidics due to the small sample volume necessary.[63] Digital microfluidic systems can be combined with a macrofluidic system designed to decrease sample volume, in turn increasing analyte concentration.[63] It follows the same principles as the magnetic particles for separation, but includes pumping of the droplet to cycle a larger volume of fluid around the magnetic particles.[63] Extraction of drug analytes from dried urine samples has also been reported. A droplet of extraction solvent, in this case methanol, is repeatedly flowed over a sample of dried urine sample then moved to a final electrode where the liquid is extracted through a capillary and then analyzed using mass spectrometry.[91]
Immunoassays
[edit]The advanced fluid handling capabilities of digital microfluidics (DMF) allows for the adoption of DMF as an immunoassay platform as DMF devices can precisely manipulate small quantities of liquid reagents. Both heterogeneous immunoassays (antigens interacting with immobilized antibodies) and homogeneous immunoassays (antigens interacting with antibodies in solution) have been developed using a DMF platform.[92] With regards to heterogeneous immunoassays, DMF can simplify the extended and intensive procedural steps by performing all delivery, mixing, incubation, and washing steps on the surface of the device (on-chip). Further, existing immunoassay techniques and methods, such as magnetic bead-based assays, ELISAs, and electrochemical detection, have been incorporated onto DMF immunoassay platforms.[93][94][95][96]
The incorporation of magnetic bead-based assays onto a DMF immunoassay platform has been demonstrated for the detection of multiple analytes, such as human insulin, IL-6, cardiac marker Troponin I (cTnI), thyroid stimulating hormone (TSH), sTNF-RI, and 17β-estradiol.[95][97][98][99] For example, a magnetic bead-based approached has been used for the detection of cTnI from whole blood in less than 8 minutes.[97] Briefly, magnetic beads containing primary antibodies were mixed with labeled secondary antibodies, incubated, and immobilized with a magnet for the washing steps. The droplet was then mixed with a chemiluminescent reagent and detection of the accompanying enzymatic reaction was measured on-chip with a photomultiplier tube.
The ELISA template, commonly used for performing immunoassays and other enzyme-based biochemical assays, has been adapted for use with the DMF platform for the detection of analytes such as IgE and IgG.[100][101] In one example,[93] a series of bioassays were conducted to establish the quantification capabilities of DMF devices, including an ELISA-based immunoassay for the detection of IgE. Superparamagnetic nanoparticles were immobilized with anti-IgE antibodies and fluorescently labeled aptamers to quantify IgE using an ELISA template. Similarly, for the detection of IgG, IgG can be immobilized onto a DMF chip, conjugated with horseradish-peroxidase (HRP)-labeled IgG, and then quantified through measurement of the color change associated with product formation of the reaction between HRP and tetramethylbenzidine.[100]
To further expand the capabilities and applications of DMF immunoassays beyond colorimetric detection (i.e., ELISA, magnetic bead-based assays), electrochemical detection tools (e.g., microelectrodes) have been incorporated into DMF chips for the detection of analytes such as TSH and rubella virus.[96][102][103] For example, Rackus et al.[102] integrated microelectrodes onto a DMF chip surface and substituted a previously reported chemiluminescent IgG immunoassay[104] with an electroactive species, enabling detection of rubella virus. They coated magnetic beads with rubella virus, anti-rubella IgG, and anti-human IgG coupled with alkaline phosphatase, which in turn catalyzed an electron transfer reaction that was detected by the on-chip microelectrodes.
Mass spectrometry
[edit]The coupling of digital microfluidics (DMF) and Mass Spectrometry can largely be categorized into indirect off-line analysis, direct off-line analysis, and in-line analysis[17] and the main advantages of this coupling are decreased solvent and reagent use, as well as decreased analysis times.[105]
Indirect off-line analysis is the usage of DMF devices to combine reactants and isolate products, which are then removed and manually transferred to a mass spectrometer. This approach takes advantage of DMF for the sample preparation step but also introduces opportunities for contamination as manual intervention is required to transfer the sample. In one example of this technique, a Grieco three-component condensation was carried out on chip and was taken off the chip by micropipette for quenching and further analysis.[77]
Direct off-line analysis is the usage of DMF devices that have been fabricated and incorporated partially or totally into a mass spectrometer. This process is still considered off-line, however as some post-reaction procedures may be carried out manually (but on chip), without the use of the digital capabilities of the device. Such devices are most often used in conjugation with MALDI-MS. In MALDI-based direct off-line devices, the droplet must be dried and recrystallized along with matrix – operations that oftentimes require vacuum chambers.[17][106] The chip with crystallized analyte is then placed in to the MALDI-MS for analysis. One issue raised with MALDI-MS coupling to DMF is that the matrix necessary for MALDI-MS can be highly acidic, which may interfere with the on-chip reactions[107]
Inline analysis is the usage of devices that feed directly into mass spectrometers, thereby eliminating any manual manipulation. Inline analysis may require specially fabricated devices and connecting hardware between the device and the mass spectrometer.[17] Inline analysis is often coupled with electrospray ionization. In one example, a DMF chip was fabricated with a hole that led to a microchannel[108] This microchannel was, in turn, connected to an electrospray ionizer that emitted directly into a mass spectrometer. Integration ambient ionization techniques where ions are formed outside of the mass spectrometer with little or no treatment pairs well with the open or semi-open microfluidic nature of DMF and allows easy inline couping between DMF and MS systems. Ambient Ionization techniques such as Surface Acoustic Wave (SAW) ionization generate surface waves on a flat piezoelectric surface that imparts enough acoustic energy on the liquid interface to overcome surface tension and desorb ions off the chip into the mass analyzer.[109][17] Some couplings utilize an external high-voltage pulse source at the physical inlet to the mass spectrometer [110] but the true role of such additions is uncertain.[111]
A significant barrier to the widespread integration of DMF with mass spectrometry is biological contamination, often termed bio-fouling.[17] High throughput analysis is a significant advantage in the use of DMF systems,[105] but means that they are particularly susceptible to cross contamination between experiments. As a result, the coupling of DMF with mass spectrometry often requires the integration of a variety of methods to prevent cross contamination such as multiple washing steps,[112][113] biologically compatible surfactants,[114] and or super hydrophobic surfaces to prevent droplet adsorption.[115][116] In one example, a reduction in cross contaminant signal during the characterization of an amino acid required 4-5 wash steps between each sample droplet for the contamination intensity to fall below the limit of detection.[113]
Miniature Mass Spectrometers
[edit]Conventional mass spectrometers are often large as well as prohibitively expensive and complex in their operation which has led to the increased attractiveness of miniature mass spectrometers (MMS) for a variety of applications. MMS are optimized towards affordability and simple operation, often forgoing the need for experienced technicians, having a low cost of manufacture, and being small enough in size to allow for the transfer of data collection from the laboratory into the field.[117] These advantages often come at the cost of reduced performance where MMS resolution, as well as the limits of detection and quantitation, are often barely adequate to perform specialized tasks. The integration of DMF with MMS has the potential for significant improvement of MMS systems by increasing throughput, resolution, and automation, while decreasing solvent cost, enabling lab grade analysis at a much reduced cost. In one example the use of a custom DMF system for urine drug testing enabled the creation of an instrument weighing only 25 kg with performance comparable to standard laboratory analysis.[118]
Nuclear magnetic resonance spectroscopy
[edit]Nuclear magnetic resonance (NMR) spectroscopy can be used in conjunction with digital microfluidics (DMF) through the use of NMR microcoils, which are electromagnetic conducting coils that are less than 1 mm in size. Due to their size, these microcoils have several limitations, directly influencing the sensitivity of the machinery they operate within.
Microchannel/microcoil interfaces, previous to digital microfluidics, had several drawbacks such as in that many created large amounts of solvent waste and were easily contaminated.[119][120] In this way, the use of digital microfluidics and its capability to manipulate singlet droplets is promising.
The interface between digital microfluidics and NMR relaxometry has led to the creation of systems such as those used to detect and quantify the concentrations of specific molecules on microscales[120] with some such systems using two step processes in which DMF devices guide droplets to the NMR detection site.[121] Introductory systems of high-field NMR and 2D NMR in conjunction with microfluidics have also been developed.[119] These systems use single plate DMF devices with NMR microcoils in place of the second plate. Recently, further modified version of this interface included pulsed field gradients (PFG) units that enabled this platform to perform more sophisticated NMR measurements (e.g. NMR diffusometry, gradients encoded pulse measurements).[122] This system has been successfully applied into monitoring rapid organic reactions.[123]
References
[edit]- ^ a b Shamsi MH, Choi K, Ng AH, Chamberlain MD, Wheeler AR (March 2016). "Electrochemiluminescence on digital microfluidics for microRNA analysis". Biosensors & Bioelectronics (Submitted manuscript). 77: 845–52. doi:10.1016/j.bios.2015.10.036. PMID 26516684.
- ^ "Duke Microfluidics Lab". microfluidics.ee.duke.edu. Retrieved 2017-05-22.
- ^ Kim CJ (November 2001). Micropumping by Electrowetting. Proc. ASME Int. Mechanical Engineering Congress and Exposition. New York, NY. IMECE2001/HTD-24200.
- ^ a b c Jain V, Devarasetty V, Patrikar R (June 2017). "Effect of electrode geometry on droplet velocity in open EWOD based device for digital microfluidics applications". Journal of Electrostatics. 87: 11–18. doi:10.1016/j.elstat.2017.02.006.
- ^ a b c d e f Choi K, Ng AH, Fobel R, Wheeler AR (2012). "Digital microfluidics". Annual Review of Analytical Chemistry. 5: 413–40. Bibcode:2012ARAC....5..413C. doi:10.1146/annurev-anchem-062011-143028. PMID 22524226.
- ^ a b c d Fair RB, Khlystov A, Tailor TD, Ivanov V, Evans RD, Srinivasan V, et al. (2007-01-01). "Chemical and Biological Applications of Digital-Microfluidic Devices". IEEE Design and Test of Computers. 24 (1): 10–24. CiteSeerX 10.1.1.559.1440. doi:10.1109/MDT.2007.8. S2CID 10122940.
- ^ Shamsi MH, Choi K, Ng AH, Chamberlain MD, Wheeler AR (March 2016). "Electrochemiluminescence on digital microfluidics for microRNA analysis". Biosensors & Bioelectronics. 77: 845–52. doi:10.1016/j.bios.2015.10.036. PMID 26516684.
- ^ Zhao Y, Xu T, Chakrabarty K (2011-07-01). "Broadcast Electrode-Addressing and Scheduling Methods for Pin-Constrained Digital Microfluidic Biochips". IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems. 30 (7): 986–999. doi:10.1109/TCAD.2011.2116250. ISSN 0278-0070. S2CID 4159209.
- ^ a b c d Berthier J (2008). Microdrops and digital microfluidics. William Andrew Pub. ISBN 9780815515449. OCLC 719878673.
- ^ Chiou PY, Moon H, Toshiyoshi H, Kim CJ, Wu MC (May 2003). "Light actuation of liquid by optoelectrowetting". Sensors and Actuators A: Physical. 104 (3): 222–8. Bibcode:2003SeAcA.104..222C. doi:10.1016/S0924-4247(03)00024-4.
- ^ Arscott S (2011). "Moving liquids with light: photoelectrowetting on semiconductors". Scientific Reports. 1: 184. arXiv:1108.4935. Bibcode:2011NatSR...1E.184A. doi:10.1038/srep00184. PMC 3240946. PMID 22355699.
- ^ Palma C, Deegan RD (March 2018). "Droplet Translation Actuated by Photoelectrowetting". Langmuir: The ACS Journal of Surfaces and Colloids. 34 (10): 3177–3185. doi:10.1021/acs.langmuir.7b03340. PMID 29457909.
- ^ Goodman J. "Water Drops: Cohesion and Adhesion of Water". www.appstate.edu. Retrieved 2017-05-21.
- ^ "Wetting". web.mit.edu. Retrieved 2017-05-21.
- ^ a b c d e f g h i j k l Cho SK, Moon H, Kim CJ (February 2003). "Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits" (PDF). Journal of Microelectromechanical Systems. 12 (1): 70–80. doi:10.1109/JMEMS.2002.807467.
- ^ Chang JH, Kim DS, Pak JJ (2011-05-02). "Simplified Ground-type Single-plate Electrowetting Device for Droplet Transport". Journal of Electrical Engineering & Technology. 6 (3): 402–407. doi:10.5370/JEET.2011.6.3.402. ISSN 1975-0102.
- ^ a b c d e f g Kirby AE, Wheeler AR (July 2013). "Digital microfluidics: an emerging sample preparation platform for mass spectrometry". Analytical Chemistry. 85 (13): 6178–84. doi:10.1021/ac401150q. PMID 23777536.
- ^ a b Teh SY, Lin R, Hung LH, Lee AP (February 2008). "Droplet microfluidics". Lab on a Chip. 8 (2): 198–220. doi:10.1039/B715524G. PMID 18231657.
- ^ a b Pollack MG, Fair RB, Shenderov AD (2000-09-11). "Electrowetting-based actuation of liquid droplets for microfluidic applications". Applied Physics Letters. 77 (11): 1725–1726. Bibcode:2000ApPhL..77.1725P. doi:10.1063/1.1308534. ISSN 0003-6951.
- ^ Nikapitiya NY, Nahar MM, Moon H (2017-06-16). "Accurate, consistent, and fast droplet splitting and dispensing in electrowetting on dielectric digital microfluidics". Micro and Nano Systems Letters. 5 (1): 24. Bibcode:2017MNSL....5...24N. doi:10.1186/s40486-017-0058-6. ISSN 2213-9621.
- ^ a b c d Banerjee A, Liu Y, Heikenfeld J, Papautsky I (December 2012). "Deterministic splitting of fluid volumes in electrowetting microfluidics". Lab on a Chip. 12 (24): 5138–5141. doi:10.1039/c2lc40723j. PMID 23042521.
- ^ a b Liu Y, Banerjee A, Papautsky I (2014-01-10). "Precise droplet volume measurement and electrode-based volume metering in digital microfluidics". Microfluidics and Nanofluidics. 17 (2): 295–303. doi:10.1007/s10404-013-1318-2. ISSN 1613-4982. S2CID 16884950.
- ^ Vergauwe N, Witters D, Atalay YT, Verbruggen B, Vermeir S, Ceyssens F, et al. (2011-01-26). "Controlling droplet size variability of a digital lab-on-a-chip for improved bio-assay performance". Microfluidics and Nanofluidics. 11 (1): 25–34. doi:10.1007/s10404-011-0769-6. ISSN 1613-4982. S2CID 93039641.
- ^ Shamsi MH, Choi K, Ng AH, Wheeler AR (February 2014). "A digital microfluidic electrochemical immunoassay". Lab on a Chip. 14 (3): 547–554. doi:10.1039/c3lc51063h. PMID 24292705.
- ^ Chang YH, Lee GB, Huang FC, Chen YY, Lin JL (September 2006). "Integrated polymerase chain reaction chips utilizing digital microfluidics". Biomedical Microdevices. 8 (3): 215–225. doi:10.1007/s10544-006-8171-y. PMID 16718406. S2CID 21275449.
- ^ Fan SK, Hashi C, Kim CJ (2003). "Manipulation of multiple droplets on N×M grid by cross-reference EWOD driving scheme and pressure-contact packaging". The Sixteenth Annual International Conference on Micro Electro Mechanical Systems, 2003. MEMS-03 Kyoto. IEEE. pp. 694–697. doi:10.1109/MEMSYS.2003.1189844. ISBN 0-7803-7744-3. S2CID 108612930.
- ^ a b Elvira KS, Leatherbarrow R, Edel J, Demello A (June 2012). "Droplet dispensing in digital microfluidic devices: Assessment of long-term reproducibility". Biomicrofluidics. 6 (2): 22003–2200310. doi:10.1063/1.3693592. PMC 3360711. PMID 22655007.
- ^ a b Nikapitiya NJ, You SM, Moon H (2014). "Droplet dispensing and splitting by electrowetting on dielectric digital microfluidics". 2014 IEEE 27th International Conference on Micro Electro Mechanical Systems (MEMS). pp. 955–958. doi:10.1109/MEMSYS.2014.6765801. ISBN 978-1-4799-3509-3. S2CID 45003766.
- ^ a b c Accardo A, Mecarini F, Leoncini M, Brandi F, Di Cola E, Burghammer M, et al. (February 2013). "Fast, active droplet interaction: coalescence and reactive mixing controlled by electrowetting on a superhydrophobic surface". Lab on a Chip. 13 (3): 332–335. doi:10.1039/c2lc41193h. PMID 23224020.
- ^ a b c Wang W, Jones TB (2011-06-23). "Microfluidic actuation of insulating liquid droplets in a parallel-plate device". Journal of Physics: Conference Series. 301 (1): 012057. Bibcode:2011JPhCS.301a2057W. doi:10.1088/1742-6596/301/1/012057. ISSN 1742-6596.
- ^ Phan SK, Hashi C, Kim CJ (2003). "Manipulation of multiple droplets on N×M grid by cross-reference EWOD driving scheme and pressure-contact packaging". The Sixteenth Annual International Conference on Micro Electro Mechanical Systems, 2003. MEMS-03 Kyoto. IEEE. IEEE. pp. 694–697. doi:10.1109/memsys.2003.1189844. ISBN 0-7803-7744-3. S2CID 108612930.
- ^ Fair RB, Khlystov A, Tailor TD, Ivanov V, Evans RD, Srinivasan V, et al. (January 2007). "Chemical and Biological Applications of Digital-Microfluidic Devices". IEEE Design & Test of Computers. 24 (1): 10–24. doi:10.1109/MDT.2007.8. hdl:10161/6987. ISSN 0740-7475. S2CID 10122940.
- ^ a b Banerjee A, Noh JH, Liu Y, Rack PD, Papautsky I (2015-01-22). "Programmable Electrowetting with Channels and Droplets". Micromachines. 6 (2): 172–185. doi:10.3390/mi6020172. ISSN 2072-666X.
- ^ a b c d Roux JM, Fouillet Y, Achard JL (March 2007). "3D droplet displacement in microfluidic systems by electrostatic actuation" (PDF). Sensors and Actuators A: Physical. 134 (2): 486–93. Bibcode:2007SeAcA.134..486R. doi:10.1016/j.sna.2006.05.012. S2CID 108644890.
- ^ Fouillet Y, Achard JL (June 2004). "Microfluidique discrète et biotechnologie" (PDF). Comptes Rendus Physique. 5 (5): 577–88. Bibcode:2004CRPhy...5..577F. doi:10.1016/j.crhy.2004.04.004.
- ^ a b Kolar P, Fair RB (2001). Non-contact electrostatic stamping for DNA microarray synthesis (poster). Proceedings of the SmallTalk2001. San Diego, USA.
- ^ a b Lebedev NN, Skal'skaya IP (1962). "Force acting on a conducting sphere in the field of a parallel plate condenser". Soviet Phys. Tech. Phys. 7: 268–270.
- ^ Velev OD, Prevo BG, Bhatt KH (December 2003). "On-chip manipulation of free droplets". Nature. 426 (6966): 515–6. Bibcode:2003Natur.426..515V. doi:10.1038/426515a. PMID 14654830. S2CID 21293602.
- ^ Gascoyne PR, Vykoukal JV, Schwartz JA, Anderson TJ, Vykoukal DM, Current KW, McConaghy C, Becker FF, Andrews C (August 2004). "Dielectrophoresis-based programmable fluidic processors". Lab on a Chip. 4 (4): 299–309. doi:10.1039/b404130e. PMID 15269795.
- ^ Taniguchi T, Torii T, Higuchi T (February 2002). "Chemical reactions in microdroplets by electrostatic manipulation of droplets in liquid media". Lab on a Chip. 2 (1): 19–23. doi:10.1039/b108739h. PMID 15100855.
- ^ Coelho B, Veigas B, Fortunato E, Martins R, Águas H, Igreja R, Baptista PV (June 2017). "Digital Microfluidics for Nucleic Acid Amplification". Sensors. 17 (7): 1495. Bibcode:2017Senso..17.1495C. doi:10.3390/s17071495. PMC 5539496. PMID 28672827.
- ^ Coelho BJ, Veigas B, Bettencourt L, Águas H, Fortunato E, Martins R, et al. (March 2022). "Digital Microfluidics-Powered Real-Time Monitoring of Isothermal DNA Amplification of Cancer Biomarker". Biosensors. 12 (4): 201. doi:10.3390/bios12040201. PMC 9028060. PMID 35448261.
- ^ a b c d e f Abdelgawad M, Freire SL, Yang H, Wheeler AR (May 2008). "All-terrain droplet actuation". Lab on a Chip. 8 (5): 672–7. doi:10.1039/b801516c. PMID 18432335.
- ^ Abdelgawad M, Wheeler AR (January 2007). "Rapid prototyping in copper substrates for digital microfluidics". Advanced Materials. 19 (1): 133–7. Bibcode:2007AdM....19..133A. doi:10.1002/adma.200601818. S2CID 53621073.
- ^ a b c George SM, Moon H (March 2015). "Digital microfluidic three-dimensional cell culture and chemical screening platform using alginate hydrogels". Biomicrofluidics. 9 (2): 024116. doi:10.1063/1.4918377. PMC 4401805. PMID 25945142.
- ^ a b c Barbulovic-Nad I, Yang H, Park PS, Wheeler AR (April 2008). "Digital microfluidics for cell-based assays". Lab on a Chip. 8 (4): 519–26. doi:10.1039/b717759c. PMID 18369505.
- ^ Sparkes A, Aubrey W, Byrne E, Clare A, Khan MN, Liakata M, et al. (January 2010). "Towards Robot Scientists for autonomous scientific discovery". Automated Experimentation. 2 (1): 1. doi:10.1186/1759-4499-2-1. PMC 2813846. PMID 20119518.
- ^ Meng F, Ellis T (October 2020). "The second decade of synthetic biology: 2010-2020". Nature Communications. 11 (1): 5174. Bibcode:2020NatCo..11.5174M. doi:10.1038/s41467-020-19092-2. PMC 7560693. PMID 33057059.
- ^ Carbonell P, Radivojevic T, García Martín H (July 2019). "Opportunities at the Intersection of Synthetic Biology, Machine Learning, and Automation". ACS Synthetic Biology. 8 (7): 1474–1477. doi:10.1021/acssynbio.8b00540. hdl:20.500.11824/998. PMID 31319671. S2CID 197664634.
- ^ a b c d Kothamachu VB, Zaini S, Muffatto F (October 2020). "Role of Digital Microfluidics in Enabling Access to Laboratory Automation and Making Biology Programmable". SLAS Technology. 25 (5): 411–426. doi:10.1177/2472630320931794. PMID 32584152. S2CID 220062017.
- ^ a b Husser MC, Vo PQ, Sinha H, Ahmadi F, Shih SC (March 2018). "An Automated Induction Microfluidics System for Synthetic Biology". ACS Synthetic Biology. 7 (3): 933–944. doi:10.1021/acssynbio.8b00025. PMID 29516725.
- ^ a b c d Ruan Q, Ruan W, Lin X, Wang Y, Zou F, Zhou L, et al. (December 2020). "Digital-WGS: Automated, highly efficient whole-genome sequencing of single cells by digital microfluidics". Science Advances. 6 (50): eabd6454. Bibcode:2020SciA....6.6454R. doi:10.1126/sciadv.abd6454. PMC 7725457. PMID 33298451.
- ^ Liu D, Yang Z, Zhang L, Wei M, Lu Y (July 2020). "Cell-free biology using remote-controlled digital microfluidics for individual droplet control". RSC Advances. 10 (45): 26972–26981. Bibcode:2020RSCAd..1026972L. doi:10.1039/d0ra04588h. PMC 9055536. PMID 35515808.
- ^ Thaitrong N, Kim H, Renzi RF, Bartsch MS, Meagher RJ, Patel KD (December 2012). "Quality control of next-generation sequencing library through an integrative digital microfluidic platform". Electrophoresis. 33 (23): 3506–3513. doi:10.1002/elps.201200441. PMID 23135807. S2CID 205802837.
- ^ Baker M (2016-05-01). "1,500 scientists lift the lid on reproducibility". Nature. 533 (7604): 452–454. Bibcode:2016Natur.533..452B. doi:10.1038/533452a. ISSN 1476-4687. PMID 27225100. S2CID 4460617.
- ^ Jessop-Fabre MM, Sonnenschein N (2019). "Improving Reproducibility in Synthetic Biology". Frontiers in Bioengineering and Biotechnology. 7: 18. doi:10.3389/fbioe.2019.00018. PMC 6378554. PMID 30805337.
- ^ Sinha H, Quach AB, Vo PQ, Shih SC (July 2018). "An automated microfluidic gene-editing platform for deciphering cancer genes". Lab on a Chip. 18 (15): 2300–2312. doi:10.1039/C8LC00470F. PMID 29989627.
- ^ a b Ng AH, Li BB, Chamberlain MD, Wheeler AR (2015-12-07). "Digital Microfluidic Cell Culture". Annual Review of Biomedical Engineering. 17 (1): 91–112. doi:10.1146/annurev-bioeng-071114-040808. PMID 26643019.
- ^ a b Liu D, Yang Z, Zhang L, Wei M, Lu Y (July 2020). "Cell-free biology using remote-controlled digital microfluidics for individual droplet control". RSC Advances. 10 (45): 26972–26981. Bibcode:2020RSCAd..1026972L. doi:10.1039/D0RA04588H. PMC 9055536. PMID 35515808.
- ^ a b Wang Y, Zhao Y, Cho SK (1 October 2007). "Efficient in-droplet separation of magnetic particles for digital microfluidics". Journal of Micromechanics and Microengineering. 17 (10): 2148–2156. Bibcode:2007JMiMi..17.2148W. doi:10.1088/0960-1317/17/10/029. S2CID 135789543.
- ^ a b Vergauwe N, Vermeir S, Wacker JB, Ceyssens F, Cornaglia M, Puers R, et al. (June 2014). "A highly efficient extraction protocol for magnetic particles on a digital microfluidic chip". Sensors and Actuators B: Chemical. 196: 282–291. Bibcode:2014SeAcB.196..282V. doi:10.1016/j.snb.2014.01.076.
- ^ a b c Seale B, Lam C, Rackus DG, Chamberlain MD, Liu C, Wheeler AR (October 2016). "Digital Microfluidics for Immunoprecipitation". Analytical Chemistry. 88 (20): 10223–10230. doi:10.1021/acs.analchem.6b02915. PMID 27700039.
- ^ a b c d e Shah GJ, Kim CJ (April 2009). "Meniscus-Assisted High-Efficiency Magnetic Collection and Separation for EWOD Droplet Microfluidics". Journal of Microelectromechanical Systems. 18 (2): 363–375. doi:10.1109/JMEMS.2009.2013394. S2CID 24845666.
- ^ a b Jebrail MJ, Sinha A, Vellucci S, Renzi RF, Ambriz C, Gondhalekar C, et al. (April 2014). "World-to-digital-microfluidic interface enabling extraction and purification of RNA from human whole blood". Analytical Chemistry. 86 (8): 3856–3862. doi:10.1021/ac404085p. PMID 24479881.
- ^ a b Hung PY, Jiang PS, Lee EF, Fan SK, Lu YW (April 2015). "Genomic DNA extraction from whole blood using a digital microfluidic (DMF) platform with magnetic beads". Microsystem Technologies. 23 (2): 313–320. doi:10.1007/s00542-015-2512-9. S2CID 137531469.
- ^ a b Choi K, Ng AH, Fobel R, Chang-Yen DA, Yarnell LE, Pearson EL, et al. (October 2013). "Automated digital microfluidic platform for magnetic-particle-based immunoassays with optimization by design of experiments". Analytical Chemistry. 85 (20): 9638–9646. doi:10.1021/ac401847x. PMID 23978190.
- ^ a b Choi K, Boyacı E, Kim J, Seale B, Barrera-Arbelaez L, Pawliszyn J, Wheeler AR (April 2016). "A digital microfluidic interface between solid-phase microextraction and liquid chromatography-mass spectrometry". Journal of Chromatography A. 1444: 1–7. doi:10.1016/j.chroma.2016.03.029. PMID 27048987.
- ^ a b Wijethunga PA, Nanayakkara YS, Kunchala P, Armstrong DW, Moon H (March 2011). "On-chip drop-to-drop liquid microextraction coupled with real-time concentration monitoring technique". Analytical Chemistry. 83 (5): 1658–1664. doi:10.1021/ac102716s. PMID 21294515.
- ^ a b Shah GJ, Ohta AT, Chiou EP, Wu MC, Kim CJ (June 2009). "EWOD-driven droplet microfluidic device integrated with optoelectronic tweezers as an automated platform for cellular isolation and analysis". Lab on a Chip. 9 (12): 1732–1739. doi:10.1039/b821508a. PMID 19495457.
- ^ a b c Nejad HR, Samiei E, Ahmadi A, Hoorfar M (2015). "Gravity-driven hydrodynamic particle separation in digital microfluidic systems". RSC Adv. 5 (45): 35966–35975. Bibcode:2015RSCAd...535966N. doi:10.1039/C5RA02068A.
- ^ Neuman KC, Block SM (September 2004). "Optical trapping". The Review of Scientific Instruments. 75 (9): 2787–809. Bibcode:2004RScI...75.2787N. doi:10.1063/1.1785844. PMC 1523313. PMID 16878180.
- ^ Geng H, Feng J, Stabryla LM, Cho SK (March 2017). "Dielectrowetting manipulation for digital microfluidics: creating, transporting, splitting, and merging of droplets". Lab on a Chip. 17 (6): 1060–1068. doi:10.1039/c7lc00006e. PMID 28217772.
- ^ a b c d e Jebrail MJ, Assem N, Mudrik JM, Dryden MD, Lin K, Yudin AK, Wheeler AR (2012-08-01). "Combinatorial Synthesis of Peptidomimetics Using Digital Microfluidics". Journal of Flow Chemistry. 2 (3): 103–107. Bibcode:2012JFlCh...2..103J. doi:10.1556/JFC-D-12-00012. S2CID 34049157.
- ^ a b Chen S, Javed MR, Kim HK, Lei J, Lazari M, Shah GJ, et al. (March 2014). "Radiolabelling diverse positron emission tomography (PET) tracers using a single digital microfluidic reactor chip". Lab on a Chip. 14 (5): 902–10. doi:10.1039/c3lc51195b. PMID 24352530. S2CID 40777981.
- ^ a b c Javed MR, Chen S, Kim HK, Wei L, Czernin J, Kim CJ, et al. (February 2014). "Efficient radiosynthesis of 3'-deoxy-3'-18F-fluorothymidine using electrowetting-on-dielectric digital microfluidic chip". Journal of Nuclear Medicine. 55 (2): 321–8. doi:10.2967/jnumed.113.121053. PMC 4494735. PMID 24365651.
- ^ Keng PY, Chen S, Ding H, Sadeghi S, Shah GJ, Dooraghi A, et al. (January 2012). "Micro-chemical synthesis of molecular probes on an electronic microfluidic device". Proceedings of the National Academy of Sciences of the United States of America. 109 (3): 690–5. Bibcode:2012PNAS..109..690K. doi:10.1073/pnas.1117566109. PMC 3271918. PMID 22210110.
- ^ a b c d Dubois P, Marchand G, Fouillet Y, Berthier J, Douki T, Hassine F, et al. (July 2006). "Ionic liquid droplet as e-microreactor". Analytical Chemistry. 78 (14): 4909–17. doi:10.1021/ac060481q. PMID 16841910.
- ^ Um T, Hong J, Im do J, Lee SJ, Kang IS (August 2016). "Electrically Controllable Microparticle Synthesis and Digital Microfluidic Manipulation by Electric-Field-Induced Droplet Dispensing into Immiscible Fluids". Scientific Reports. 6 (1): 31901. Bibcode:2016NatSR...631901U. doi:10.1038/srep31901. PMC 4989170. PMID 27534580.
- ^ a b Witters D, Vergauwe N, Ameloot R, Vermeir S, De Vos D, Puers R, et al. (March 2012). "Digital microfluidic high-throughput printing of single metal-organic framework crystals". Advanced Materials. 24 (10): 1316–20. Bibcode:2012AdM....24.1316W. doi:10.1002/adma.201104922. PMID 22298246. S2CID 205244275.
- ^ a b c Jebrail MJ, Ng AH, Rai V, Hili R, Yudin AK, Wheeler AR (November 2010). "Synchronized synthesis of peptide-based macrocycles by digital microfluidics". Angewandte Chemie. 49 (46): 8625–8629. doi:10.1002/anie.201001604. PMID 20715231.
- ^ Luk VN, Wheeler AR (June 2009). "A digital microfluidic approach to proteomic sample processing". Analytical Chemistry. 81 (11): 4524–4530. doi:10.1021/ac900522a. hdl:1807/34790. PMID 19476392.
- ^ Sadeghi S, Ding H, Shah GJ, Chen S, Keng PY, Kim CJ, van Dam RM (February 2012). "On chip droplet characterization: a practical, high-sensitivity measurement of droplet impedance in digital microfluidics". Analytical Chemistry. 84 (4): 1915–1923. doi:10.1021/ac202715f. PMID 22248060. S2CID 9113055.
- ^ Wu B, von der Ecken S, Swyer I, Li C, Jenne A, Vincent F, et al. (October 2019). "Rapid Chemical Reaction Monitoring by Digital Microfluidics-NMR: Proof of Principle Towards an Automated Synthetic Discovery Platform". Angewandte Chemie. 58 (43): 15372–15376. doi:10.1002/anie.201910052. PMID 31449724. S2CID 201728604.
- ^ Peck TL, Magin RL, Lauterbur PC (August 1995). "Design and analysis of microcoils for NMR microscopy". Journal of Magnetic Resonance, Series B. 108 (2): 114–124. Bibcode:1995JMRB..108..114P. doi:10.1006/jmrb.1995.1112. PMID 7648010.
- ^ a b Moazami E, Perry JM, Soffer G, Husser MC, Shih SC (April 2019). "Integration of World-to-Chip Interfaces with Digital Microfluidics for Bacterial Transformation and Enzymatic Assays". Analytical Chemistry. 91 (8): 5159–5168. doi:10.1021/acs.analchem.8b05754. PMID 30945840. S2CID 93000574.
- ^ a b Ng AH, Dean Chamberlain M, Situ H, Lee V, Wheeler AR (June 2015). "Digital microfluidic immunocytochemistry in single cells". Nature Communications. 6 (1): 7513. Bibcode:2015NatCo...6.7513N. doi:10.1038/ncomms8513. PMC 4491823. PMID 26104298.
- ^ a b Aijian AP, Garrell RL (June 2015). "Digital microfluidics for automated hanging drop cell spheroid culture". Journal of Laboratory Automation. 20 (3): 283–95. doi:10.1177/2211068214562002. PMID 25510471. S2CID 23720265.
- ^ a b Ben Yehezkel T, Rival A, Raz O, Cohen R, Marx Z, Camara M, et al. (February 2016). "Synthesis and cell-free cloning of DNA libraries using programmable microfluidics". Nucleic Acids Research. 44 (4): e35. doi:10.1093/nar/gkv1087. PMC 4770201. PMID 26481354.
- ^ Fan SK, Hsu YW, Chen CH (August 2011). "Encapsulated droplets with metered and removable oil shells by electrowetting and dielectrophoresis". Lab on a Chip. 11 (15): 2500–8. doi:10.1039/c1lc20142e. PMID 21666906.
- ^ "Millipore and HyClone form bioprocessing alliance". Membrane Technology. 2004 (3): 1. March 2004. doi:10.1016/s0958-2118(04)00087-4. ISSN 0958-2118.
- ^ Kirby AE, Lafrenière NM, Seale B, Hendricks PI, Cooks RG, Wheeler AR (June 2014). "Analysis on the go: quantitation of drugs of abuse in dried urine with digital microfluidics and miniature mass spectrometry". Analytical Chemistry. 86 (12): 6121–9. doi:10.1021/ac5012969. PMID 24906177.
- ^ Ng AH, Uddayasankar U, Wheeler AR (June 2010). "Immunoassays in microfluidic systems. Analytical and bioanalytical chemistry". Analytical and Bioanalytical Chemistry. 397 (3): 991–1007. doi:10.1007/s00216-010-3678-8. PMID 20422163. S2CID 30670634.
- ^ a b Vergauwe N, Witters D, Ceyssens F, Vermeir S, Verbruggen B, Puers R, Lammertyn J (April 2011). "A versatile electrowetting-based digital microfluidic platform for quantitative homogeneous and heterogeneous bio-assays". Journal of Micromechanics and Microengineering. 21 (5): 054026. Bibcode:2011JMiMi..21e4026V. doi:10.1088/0960-1317/21/5/054026. S2CID 111122895.
- ^ Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack M, Pamula V (December 2008). "Development of a digital microfluidic platform for point of care testing". Lab on a Chip. 8 (12): 2091–104. doi:10.1039/b814922d. PMC 2726010. PMID 19023472.
- ^ a b Ng AH, Choi K, Luoma RP, Robinson JM, Wheeler AR (October 2012). "Digital microfluidic magnetic separation for particle-based immunoassays". Analytical Chemistry. 84 (20): 8805–12. doi:10.1021/ac3020627. PMID 23013543.
- ^ a b Shamsi MH, Choi K, Ng AH, Wheeler AR (February 2014). "A digital microfluidic electrochemical immunoassay". Lab on a Chip. 14 (3): 547–54. doi:10.1039/c3lc51063h. PMID 24292705.
- ^ a b Sista RS, Eckhardt AE, Srinivasan V, Pollack MG, Palanki S, Pamula VK (December 2008). "Heterogeneous immunoassays using magnetic beads on a digital microfluidic platform". Lab on a Chip. 8 (12): 2188–96. doi:10.1039/b807855f. PMC 2726047. PMID 19023486.
- ^ Tsaloglou MN, Jacobs A, Morgan H (September 2014). "A fluorogenic heterogeneous immunoassay for cardiac muscle troponin cTnI on a digital microfluidic device". Analytical and Bioanalytical Chemistry. 406 (24): 5967–76. doi:10.1007/s00216-014-7997-z. PMID 25074544. S2CID 24266593.
- ^ Huang CY, Tsai PY, Lee IC, Hsu HY, Huang HY, Fan SK, Yao DJ, Liu CH, Hsu W (January 2016). "A highly efficient bead extraction technique with low bead number for digital microfluidic immunoassay". Biomicrofluidics. 10 (1): 011901. doi:10.1063/1.4939942. PMC 4714987. PMID 26858807.
- ^ a b Zhu L, Feng Y, Ye X, Feng J, Wu Y, Zhou Z (September 2012). "An ELISA chip based on an EWOD microfluidic platform". Journal of Adhesion Science and Technology. 26 (12–17): 2113–24. doi:10.1163/156856111x600172. S2CID 136668522.
- ^ Miller EM, Ng AH, Uddayasankar U, Wheeler AR (January 2011). "A digital microfluidic approach to heterogeneous immunoassays". Analytical and Bioanalytical Chemistry. 399 (1): 337–45. doi:10.1007/s00216-010-4368-2. PMID 21057776. S2CID 2809777.
- ^ a b Rackus DG, Dryden MD, Lamanna J, Zaragoza A, Lam B, Kelley SO, Wheeler AR (2015). "A digital microfluidic device with integrated nanostructured microelectrodes for electrochemical immunoassays". Lab on a Chip. 15 (18): 3776–84. doi:10.1039/c5lc00660k. PMID 26247922.
- ^ Dixon C, Ng AH, Fobel R, Miltenburg MB, Wheeler AR (November 2016). "An inkjet printed, roll-coated digital microfluidic device for inexpensive, miniaturized diagnostic assays" (PDF). Lab on a Chip. 16 (23): 4560–4568. doi:10.1039/c6lc01064d. PMID 27801455.
- ^ Ng AH, Lee M, Choi K, Fischer AT, Robinson JM, Wheeler AR (February 2015). "Digital microfluidic platform for the detection of rubella infection and immunity: a proof of concept". Clinical Chemistry. 61 (2): 420–9. doi:10.1373/clinchem.2014.232181. PMID 25512641.
- ^ a b Wang X, Yi L, Mukhitov N, Schrell AM, Dhumpa R, Roper MG (February 2015). "Microfluidics-to-mass spectrometry: a review of coupling methods and applications". Journal of Chromatography A. Editors' Choice IX. 1382: 98–116. doi:10.1016/j.chroma.2014.10.039. PMC 4318794. PMID 25458901.
- ^ Chatterjee D, Ytterberg AJ, Son SU, Loo JA, Garrell RL (March 2010). "Integration of protein processing steps on a droplet microfluidics platform for MALDI-MS analysis". Analytical Chemistry. 82 (5): 2095–101. doi:10.1021/ac9029373. PMID 20146460.
- ^ Küster SK, Fagerer SR, Verboket PE, Eyer K, Jefimovs K, Zenobi R, Dittrich PS (February 2013). "Interfacing droplet microfluidics with matrix-assisted laser desorption/ionization mass spectrometry: label-free content analysis of single droplets". Analytical Chemistry. 85 (3): 1285–9. doi:10.1021/ac3033189. PMID 23289755.
- ^ Jebrail MJ, Yang H, Mudrik JM, Lafrenière NM, McRoberts C, Al-Dirbashi OY, et al. (October 2011). "A digital microfluidic method for dried blood spot analysis". Lab on a Chip. 11 (19): 3218–24. doi:10.1039/c1lc20524b. PMID 21869989.
- ^ Yeo LY, Friend JR (January 2009). "Ultrafast microfluidics using surface acoustic waves". Biomicrofluidics. 3 (1): 12002. doi:10.1063/1.3056040. PMC 2717600. PMID 19693383.
- ^ Heron SR, Wilson R, Shaffer SA, Goodlett DR, Cooper JM (May 2010). "Surface acoustic wave nebulization of peptides as a microfluidic interface for mass spectrometry". Analytical Chemistry. 82 (10): 3985–9. doi:10.1021/ac100372c. PMC 3073871. PMID 20364823.
- ^ Ho J, Tan MK, Go DB, Yeo LY, Friend JR, Chang HC (May 2011). "Paper-based microfluidic surface acoustic wave sample delivery and ionization source for rapid and sensitive ambient mass spectrometry". Analytical Chemistry. 83 (9): 3260–6. doi:10.1021/ac200380q. PMID 21456580.
- ^ Zhao Y, Chakrabarty K (June 2010). "Synchronization of washing operations with droplet routing for cross-contamination avoidance in digital microfluidic biochips". Design Automation Conference: 635–640.
- ^ a b Shih SC, Yang H, Jebrail MJ, Fobel R, McIntosh N, Al-Dirbashi OY, et al. (April 2012). "Dried blood spot analysis by digital microfluidics coupled to nanoelectrospray ionization mass spectrometry". Analytical Chemistry. 84 (8): 3731–3738. doi:10.1021/ac300305s. PMID 22413743.
- ^ Aijian AP, Chatterjee D, Garrell RL (July 2012). "Fluorinated liquid-enabled protein handling and surfactant-aided crystallization for fully in situ digital microfluidic MALDI-MS analysis". Lab on a Chip. 12 (14): 2552–2559. doi:10.1039/C2LC21135A. PMID 22569918.
- ^ Samiei E, Tabrizian M, Hoorfar M (July 2016). "A review of digital microfluidics as portable platforms for lab-on a-chip applications". Lab on a Chip. 16 (13): 2376–2396. doi:10.1039/C6LC00387G. PMID 27272540.
- ^ Lapierre F, Piret G, Drobecq H, Melnyk O, Coffinier Y, Thomy V, Boukherroub R (May 2011). "High sensitive matrix-free mass spectrometry analysis of peptides using silicon nanowires-based digital microfluidic device". Lab on a Chip. 11 (9): 1620–1628. doi:10.1039/C0LC00716A. PMID 21423926.
- ^ Ouyang Z, Cooks RG (2009-07-19). "Miniature mass spectrometers". Annual Review of Analytical Chemistry. 2 (1): 187–214. Bibcode:2009ARAC....2..187O. doi:10.1146/annurev-anchem-060908-155229. PMID 20636059.
- ^ Kirby AE, Lafrenière NM, Seale B, Hendricks PI, Cooks RG, Wheeler AR (June 2014). "Analysis on the go: quantitation of drugs of abuse in dried urine with digital microfluidics and miniature mass spectrometry". Analytical Chemistry. 86 (12): 6121–6129. doi:10.1021/ac5012969. PMID 24906177.
- ^ a b Swyer I, Soong R, Dryden MD, Fey M, Maas WE, Simpson A, Wheeler AR (November 2016). "Interfacing digital microfluidics with high-field nuclear magnetic resonance spectroscopy". Lab on a Chip. 16 (22): 4424–4435. doi:10.1039/c6lc01073c. PMID 27757467.
- ^ a b Lei KM, Mak PI, Law MK, Martins RP (August 2015). "A palm-size μNMR relaxometer using a digital microfluidic device and a semiconductor transceiver for chemical/biological diagnosis". The Analyst. 140 (15): 5129–37. Bibcode:2015Ana...140.5129L. doi:10.1039/c5an00500k. PMID 26034784.
- ^ Lei KM, Mak PI, Law MK, Martins RP (December 2014). "NMR-DMF: a modular nuclear magnetic resonance-digital microfluidics system for biological assays". The Analyst. 139 (23): 6204–13. Bibcode:2014Ana...139.6204L. doi:10.1039/c4an01285b. PMID 25315808.
- ^ Swyer I, von der Ecken S, Wu B, Jenne A, Soong R, Vincent F, Schmidig D, Frei T, Busse F, Stronks HJ, Simpson AJ, Wheeler AR (January 2019). "Digital microfluidics and nuclear magnetic resonance spectroscopy for in situ diffusion measurements and reaction monitoring". Lab on a Chip. 19 (4): 641–653. doi:10.1039/C8LC01214H. PMID 30648175. S2CID 58600090.
- ^ Wu B, von der Ecken S, Swyer I, Li CL, Jenne A, Vincent F, Schmidig D, Kuehn T, Beck A, Busse F, Stronks HJ, Soong R, Wheeler AR, Simpson AJ (October 2019). "Rapid Chemical Reaction Monitoring by Digital Microfluidics-NMR: Proof of Principle Towards an Automated Synthetic Discovery Platform". Angewandte Chemie International Edition. 58 (43): 15372–15376. doi:10.1002/anie.201910052. PMID 31449724. S2CID 201728604.











